Tools
Software Defects Prediction using Machine Learning
2025-12-31
0 views
admin
This Python 3 environment comes with many helpful analytics libraries installed ## It is defined by the kaggle/python Docker image: https://github.com/kaggle/docker-python ## For example, here's several helpful packages to load Step 1: Data Loading and Initial Analysis Handling Missing Values Step 4 : Model Building and Prediction Templates let you quickly answer FAQs or store snippets for re-use. Are you sure you want to hide this comment? It will become hidden in your post, but will still be visible via the comment's permalink. Hide child comments as well For further actions, you may consider blocking this person and/or reporting abuse COMMAND_BLOCK:
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.preprocessing import StandardScaler
import warnings
warnings.filterwarnings('ignore') # Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory import os
for dirname, _, filenames in os.walk('/kaggle/input'): for filename in filenames: print(os.path.join(dirname, filename)) # You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All" # You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.preprocessing import StandardScaler
import warnings
warnings.filterwarnings('ignore') # Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory import os
for dirname, _, filenames in os.walk('/kaggle/input'): for filename in filenames: print(os.path.join(dirname, filename)) # You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All" # You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session COMMAND_BLOCK:
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.preprocessing import StandardScaler
import warnings
warnings.filterwarnings('ignore') # Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory import os
for dirname, _, filenames in os.walk('/kaggle/input'): for filename in filenames: print(os.path.join(dirname, filename)) # You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All" # You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session COMMAND_BLOCK:
# Load the Defects Dataset
train = pd.read_csv('/kaggle/input/train.csv')
test = pd.read_csv('/kaggle/input/test.csv') # Display first few rows
print("First 5 rows of the training dataset:")
print(train.head())
print("First 5 rows of the test dataset:")
print(test.head()) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Load the Defects Dataset
train = pd.read_csv('/kaggle/input/train.csv')
test = pd.read_csv('/kaggle/input/test.csv') # Display first few rows
print("First 5 rows of the training dataset:")
print(train.head())
print("First 5 rows of the test dataset:")
print(test.head()) COMMAND_BLOCK:
# Load the Defects Dataset
train = pd.read_csv('/kaggle/input/train.csv')
test = pd.read_csv('/kaggle/input/test.csv') # Display first few rows
print("First 5 rows of the training dataset:")
print(train.head())
print("First 5 rows of the test dataset:")
print(test.head()) COMMAND_BLOCK:
# Dataset shape
print(f"\nDataset Shape: {train.shape}")
print(f"Number of samples: {train.shape[0]}")
print(f"Number of features: {train.shape[1]}") print(f"\nDataset Shape: {test.shape}")
print(f"Number of samples: {test.shape[0]}")
print(f"Number of features: {test.shape[1]}") Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Dataset shape
print(f"\nDataset Shape: {train.shape}")
print(f"Number of samples: {train.shape[0]}")
print(f"Number of features: {train.shape[1]}") print(f"\nDataset Shape: {test.shape}")
print(f"Number of samples: {test.shape[0]}")
print(f"Number of features: {test.shape[1]}") COMMAND_BLOCK:
# Dataset shape
print(f"\nDataset Shape: {train.shape}")
print(f"Number of samples: {train.shape[0]}")
print(f"Number of features: {train.shape[1]}") print(f"\nDataset Shape: {test.shape}")
print(f"Number of samples: {test.shape[0]}")
print(f"Number of features: {test.shape[1]}") COMMAND_BLOCK:
# Column names and data types
print("\nColumn Information for training dataset:")
print(train.info()) print("\nColumn Information for test datase
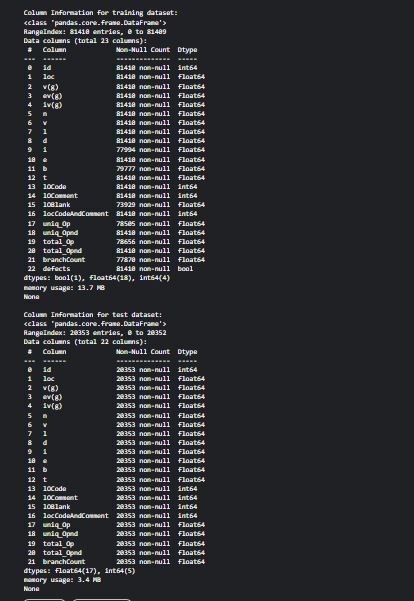t:")
print(test.info()) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Column names and data types
print("\nColumn Information for training dataset:")
print(train.info()) print("\nColumn Information for test datase
t:")
print(test.info()) COMMAND_BLOCK:
# Column names and data types
print("\nColumn Information for training dataset:")
print(train.info()) print("\nColumn Information for test datase
t:")
print(test.info()) COMMAND_BLOCK:
# Statistical summary
print("Statistical Summary:")
print(train.describe()) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Statistical summary
print("Statistical Summary:")
print(train.describe()) COMMAND_BLOCK:
# Statistical summary
print("Statistical Summary:")
print(train.describe()) CODE_BLOCK:
#Get the statistical information about the training set
import plotly.express as px
train.describe().T\ .style.bar(subset=['mean'], color=px.colors.qualitative.G10[2])\ .background_gradient(subset=['std'], cmap='Blues')\ .background_gradient(subset=['50%'], cmap='Reds') Enter fullscreen mode Exit fullscreen mode CODE_BLOCK:
#Get the statistical information about the training set
import plotly.express as px
train.describe().T\ .style.bar(subset=['mean'], color=px.colors.qualitative.G10[2])\ .background_gradient(subset=['std'], cmap='Blues')\ .background_gradient(subset=['50%'], cmap='Reds') CODE_BLOCK:
#Get the statistical information about the training set
import plotly.express as px
train.describe().T\ .style.bar(subset=['mean'], color=px.colors.qualitative.G10[2])\ .background_gradient(subset=['std'], cmap='Blues')\ .background_gradient(subset=['50%'], cmap='Reds') COMMAND_BLOCK:
# Check for missing values
print("=" * 50)
print("Missing Values Analysis")
print("=" * 50)
missing = train.isnull().sum()
missing_pct = (train.isnull().sum() / len(train)) * 100
missing_train = pd.DataFrame({ 'Missing_Count': missing, 'Missing_Percentage': missing_pct
})
print(missing_train[missing_train['Missing_Count'] > 0]) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Check for missing values
print("=" * 50)
print("Missing Values Analysis")
print("=" * 50)
missing = train.isnull().sum()
missing_pct = (train.isnull().sum() / len(train)) * 100
missing_train = pd.DataFrame({ 'Missing_Count': missing, 'Missing_Percentage': missing_pct
})
print(missing_train[missing_train['Missing_Count'] > 0]) COMMAND_BLOCK:
# Check for missing values
print("=" * 50)
print("Missing Values Analysis")
print("=" * 50)
missing = train.isnull().sum()
missing_pct = (train.isnull().sum() / len(train)) * 100
missing_train = pd.DataFrame({ 'Missing_Count': missing, 'Missing_Percentage': missing_pct
})
print(missing_train[missing_train['Missing_Count'] > 0]) COMMAND_BLOCK:
# Check for duplicates
print("=" * 50)
print("Duplicate Analysis")
print("=" * 50)
duplicates = train.duplicated().sum()
print(f"Number of duplicate rows: {duplicates}")
print(f"\nDuplicate rows:")
train[train.duplicated(keep=False)].sort_values('id') Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Check for duplicates
print("=" * 50)
print("Duplicate Analysis")
print("=" * 50)
duplicates = train.duplicated().sum()
print(f"Number of duplicate rows: {duplicates}")
print(f"\nDuplicate rows:")
train[train.duplicated(keep=False)].sort_values('id') COMMAND_BLOCK:
# Check for duplicates
print("=" * 50)
print("Duplicate Analysis")
print("=" * 50)
duplicates = train.duplicated().sum()
print(f"Number of duplicate rows: {duplicates}")
print(f"\nDuplicate rows:")
train[train.duplicated(keep=False)].sort_values('id') COMMAND_BLOCK:
# Check target variable distribution
print("\nTarget Variable Distribution:")
print(train['defects'].value_counts())
print(f"\nClass Balance:")
print(train['defects'].value_counts(normalize=True)) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Check target variable distribution
print("\nTarget Variable Distribution:")
print(train['defects'].value_counts())
print(f"\nClass Balance:")
print(train['defects'].value_counts(normalize=True)) COMMAND_BLOCK:
# Check target variable distribution
print("\nTarget Variable Distribution:")
print(train['defects'].value_counts())
print(f"\nClass Balance:")
print(train['defects'].value_counts(normalize=True)) COMMAND_BLOCK:
# target value count
defects_count = dict(train['defects'].value_counts())
defects_count Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# target value count
defects_count = dict(train['defects'].value_counts())
defects_count COMMAND_BLOCK:
# target value count
defects_count = dict(train['defects'].value_counts())
defects_count CODE_BLOCK:
#Check for missing (NaN) value
print(train[train.isna().any(axis=1)]) Enter fullscreen mode Exit fullscreen mode CODE_BLOCK:
#Check for missing (NaN) value
print(train[train.isna().any(axis=1)]) CODE_BLOCK:
#Check for missing (NaN) value
print(train[train.isna().any(axis=1)]) COMMAND_BLOCK:
# visualize data distrubution
fig , axis = plt.subplots(figsize = (25,35))
fig.suptitle('Data Distribution', ha = 'center', fontsize = 20, fontweight = 'bold',y=0.90 )
for idx,col in enumerate(list(train.columns)[1:-1]): plt.subplot(7,3,idx+1) ax = sns.histplot(train[col], color = 'blue' ,stat='density' , kde = False,bins = 50) sns.kdeplot(train[col] , color = 'orange' , ax = ax) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# visualize data distrubution
fig , axis = plt.subplots(figsize = (25,35))
fig.suptitle('Data Distribution', ha = 'center', fontsize = 20, fontweight = 'bold',y=0.90 )
for idx,col in enumerate(list(train.columns)[1:-1]): plt.subplot(7,3,idx+1) ax = sns.histplot(train[col], color = 'blue' ,stat='density' , kde = False,bins = 50) sns.kdeplot(train[col] , color = 'orange' , ax = ax) COMMAND_BLOCK:
# visualize data distrubution
fig , axis = plt.subplots(figsize = (25,35))
fig.suptitle('Data Distribution', ha = 'center', fontsize = 20, fontweight = 'bold',y=0.90 )
for idx,col in enumerate(list(train.columns)[1:-1]): plt.subplot(7,3,idx+1) ax = sns.histplot(train[col], color = 'blue' ,stat='density' , kde = False,bins = 50) sns.kdeplot(train[col] , color = 'orange' , ax = ax) COMMAND_BLOCK:
# adjust label names
labels = ['not_defects', 'defects']
# visualize data target
plt.title('Defects Values')
sns.barplot(x = labels ,y = list(defects_count.values()), width = 0.3 , palette = ['blue' , 'orange']) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# adjust label names
labels = ['not_defects', 'defects']
# visualize data target
plt.title('Defects Values')
sns.barplot(x = labels ,y = list(defects_count.values()), width = 0.3 , palette = ['blue' , 'orange']) COMMAND_BLOCK:
# adjust label names
labels = ['not_defects', 'defects']
# visualize data target
plt.title('Defects Values')
sns.barplot(x = labels ,y = list(defects_count.values()), width = 0.3 , palette = ['blue' , 'orange']) COMMAND_BLOCK:
# visualize correlation
corr = train.corr()
mask = np.triu(np.ones_like(corr))
plt.figure(figsize = (20,10))
plt.title('Correlation Matrix')
sns.heatmap(data = corr , mask = mask , annot = True , cmap = 'coolwarm',annot_kws={"color": "black", "size":9} ) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# visualize correlation
corr = train.corr()
mask = np.triu(np.ones_like(corr))
plt.figure(figsize = (20,10))
plt.title('Correlation Matrix')
sns.heatmap(data = corr , mask = mask , annot = True , cmap = 'coolwarm',annot_kws={"color": "black", "size":9} ) COMMAND_BLOCK:
# visualize correlation
corr = train.corr()
mask = np.triu(np.ones_like(corr))
plt.figure(figsize = (20,10))
plt.title('Correlation Matrix')
sns.heatmap(data = corr , mask = mask , annot = True , cmap = 'coolwarm',annot_kws={"color": "black", "size":9} ) COMMAND_BLOCK:
**Step 3 : Data cleaning and preparation**
# convert target to binary encoder
# Encode categorical variables
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
train['defects'] = le.fit_transform(train['defects']) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
**Step 3 : Data cleaning and preparation**
# convert target to binary encoder
# Encode categorical variables
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
train['defects'] = le.fit_transform(train['defects']) COMMAND_BLOCK:
**Step 3 : Data cleaning and preparation**
# convert target to binary encoder
# Encode categorical variables
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
train['defects'] = le.fit_transform(train['defects']) CODE_BLOCK:
train.defects.unique() Enter fullscreen mode Exit fullscreen mode CODE_BLOCK:
train.defects.unique() CODE_BLOCK:
train.defects.unique() COMMAND_BLOCK:
# Strategy 1: Fill missing values with mean/median
train_cleaned = train.copy() print("Before filling missing values:")
print(train_cleaned[['i', 'b','lOBlank','uniq_Op','total_Op','branchCount']].isnull().sum()) # Fill numeric missing values with median
#train_cleaned['score'].fillna(train_cleaned['score'].median(), inplace=True)
train_cleaned['i'].fillna(train_cleaned['i'].mean(), inplace=True)
train_cleaned['b'].fillna(train_cleaned['b'].mean(), inplace=True)
train_cleaned['lOBlank'].fillna(train_cleaned['lOBlank'].mean(), inplace=True)
train_cleaned['uniq_Op'].fillna(train_cleaned['uniq_Op'].mean(), inplace=True)
train_cleaned['total_Op'].fillna(train_cleaned['total_Op'].mean(), inplace=True)
train_cleaned['branchCount'].fillna(train_cleaned['branchCount'].mean(), inplace=True) print("\nAfter filling missing values:")
print(train_cleaned[['i', 'b','lOBlank','uniq_Op','total_Op','branchCount']].isnull().sum()) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Strategy 1: Fill missing values with mean/median
train_cleaned = train.copy() print("Before filling missing values:")
print(train_cleaned[['i', 'b','lOBlank','uniq_Op','total_Op','branchCount']].isnull().sum()) # Fill numeric missing values with median
#train_cleaned['score'].fillna(train_cleaned['score'].median(), inplace=True)
train_cleaned['i'].fillna(train_cleaned['i'].mean(), inplace=True)
train_cleaned['b'].fillna(train_cleaned['b'].mean(), inplace=True)
train_cleaned['lOBlank'].fillna(train_cleaned['lOBlank'].mean(), inplace=True)
train_cleaned['uniq_Op'].fillna(train_cleaned['uniq_Op'].mean(), inplace=True)
train_cleaned['total_Op'].fillna(train_cleaned['total_Op'].mean(), inplace=True)
train_cleaned['branchCount'].fillna(train_cleaned['branchCount'].mean(), inplace=True) print("\nAfter filling missing values:")
print(train_cleaned[['i', 'b','lOBlank','uniq_Op','total_Op','branchCount']].isnull().sum()) COMMAND_BLOCK:
# Strategy 1: Fill missing values with mean/median
train_cleaned = train.copy() print("Before filling missing values:")
print(train_cleaned[['i', 'b','lOBlank','uniq_Op','total_Op','branchCount']].isnull().sum()) # Fill numeric missing values with median
#train_cleaned['score'].fillna(train_cleaned['score'].median(), inplace=True)
train_cleaned['i'].fillna(train_cleaned['i'].mean(), inplace=True)
train_cleaned['b'].fillna(train_cleaned['b'].mean(), inplace=True)
train_cleaned['lOBlank'].fillna(train_cleaned['lOBlank'].mean(), inplace=True)
train_cleaned['uniq_Op'].fillna(train_cleaned['uniq_Op'].mean(), inplace=True)
train_cleaned['total_Op'].fillna(train_cleaned['total_Op'].mean(), inplace=True)
train_cleaned['branchCount'].fillna(train_cleaned['branchCount'].mean(), inplace=True) print("\nAfter filling missing values:")
print(train_cleaned[['i', 'b','lOBlank','uniq_Op','total_Op','branchCount']].isnull().sum()) COMMAND_BLOCK:
**Handling Outliers** # Visualize outliers using box plots
plt.figure(figsize=(12,12))
i=1
for col in train.columns: plt.subplot(6,6,i) train[[col]].boxplot() i+=1 Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
**Handling Outliers** # Visualize outliers using box plots
plt.figure(figsize=(12,12))
i=1
for col in train.columns: plt.subplot(6,6,i) train[[col]].boxplot() i+=1 COMMAND_BLOCK:
**Handling Outliers** # Visualize outliers using box plots
plt.figure(figsize=(12,12))
i=1
for col in train.columns: plt.subplot(6,6,i) train[[col]].boxplot() i+=1 COMMAND_BLOCK:
# Detect outliers using IQR method
def detect_outliers_iqr(data, column): Q1 = data[column].quantile(0.25) Q3 = data[column].quantile(0.75) IQR = Q3 - Q1 lower_bound = Q1 - 1.5 * IQR upper_bound = Q3 + 1.5 * IQR outliers = data[(data[column] < lower_bound) | (data[column] > upper_bound)] return outliers, lower_bound, upper_bound # Detect outliers in age
outliers_b, lower_b, upper_b = detect_outliers_iqr(train_cleaned, 'b')
print(f"b outliers (outside {lower_b:.2f} - {upper_b:.2f}):")
print(outliers_b[['id', 'b']]) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Detect outliers using IQR method
def detect_outliers_iqr(data, column): Q1 = data[column].quantile(0.25) Q3 = data[column].quantile(0.75) IQR = Q3 - Q1 lower_bound = Q1 - 1.5 * IQR upper_bound = Q3 + 1.5 * IQR outliers = data[(data[column] < lower_bound) | (data[column] > upper_bound)] return outliers, lower_bound, upper_bound # Detect outliers in age
outliers_b, lower_b, upper_b = detect_outliers_iqr(train_cleaned, 'b')
print(f"b outliers (outside {lower_b:.2f} - {upper_b:.2f}):")
print(outliers_b[['id', 'b']]) COMMAND_BLOCK:
# Detect outliers using IQR method
def detect_outliers_iqr(data, column): Q1 = data[column].quantile(0.25) Q3 = data[column].quantile(0.75) IQR = Q3 - Q1 lower_bound = Q1 - 1.5 * IQR upper_bound = Q3 + 1.5 * IQR outliers = data[(data[column] < lower_bound) | (data[column] > upper_bound)] return outliers, lower_bound, upper_bound # Detect outliers in age
outliers_b, lower_b, upper_b = detect_outliers_iqr(train_cleaned, 'b')
print(f"b outliers (outside {lower_b:.2f} - {upper_b:.2f}):")
print(outliers_b[['id', 'b']]) CODE_BLOCK:
#List of numerical columns
num_cols=[col for col in train.columns if (train[col].dtype in ["int64","float64"]) & (train[col].nunique()>50)]
num_cols Enter fullscreen mode Exit fullscreen mode CODE_BLOCK:
#List of numerical columns
num_cols=[col for col in train.columns if (train[col].dtype in ["int64","float64"]) & (train[col].nunique()>50)]
num_cols CODE_BLOCK:
#List of numerical columns
num_cols=[col for col in train.columns if (train[col].dtype in ["int64","float64"]) & (train[col].nunique()>50)]
num_cols COMMAND_BLOCK:
#Method to handle outliers
def handle_outliers( train, columns, factor=1.5, method="clip" # "clip" or "remove"
): """ Handle outliers using the IQR method. Parameters: train (pd.DataFrame): Input DataFrame columns (list): List of numeric columns factor (float): IQR multiplier (default 1.5) method (str): "clip" to cap values, "remove" to drop rows Returns: pd.DataFrame """ train = train.copy() for col in columns: Q1 = train[col].quantile(0.25) Q3 = train[col].quantile(0.75) IQR = Q3 - Q1 lower = Q1 - factor * IQR upper = Q3 + factor * IQR if method == "clip": train[col] = train[col].clip(lower, upper) elif method == "remove": train = train[(train[col] >= lower) & (train[col] <= upper)] else: raise ValueError("method must be 'clip' or 'remove'") return train Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
#Method to handle outliers
def handle_outliers( train, columns, factor=1.5, method="clip" # "clip" or "remove"
): """ Handle outliers using the IQR method. Parameters: train (pd.DataFrame): Input DataFrame columns (list): List of numeric columns factor (float): IQR multiplier (default 1.5) method (str): "clip" to cap values, "remove" to drop rows Returns: pd.DataFrame """ train = train.copy() for col in columns: Q1 = train[col].quantile(0.25) Q3 = train[col].quantile(0.75) IQR = Q3 - Q1 lower = Q1 - factor * IQR upper = Q3 + factor * IQR if method == "clip": train[col] = train[col].clip(lower, upper) elif method == "remove": train = train[(train[col] >= lower) & (train[col] <= upper)] else: raise ValueError("method must be 'clip' or 'remove'") return train COMMAND_BLOCK:
#Method to handle outliers
def handle_outliers( train, columns, factor=1.5, method="clip" # "clip" or "remove"
): """ Handle outliers using the IQR method. Parameters: train (pd.DataFrame): Input DataFrame columns (list): List of numeric columns factor (float): IQR multiplier (default 1.5) method (str): "clip" to cap values, "remove" to drop rows Returns: pd.DataFrame """ train = train.copy() for col in columns: Q1 = train[col].quantile(0.25) Q3 = train[col].quantile(0.75) IQR = Q3 - Q1 lower = Q1 - factor * IQR upper = Q3 + factor * IQR if method == "clip": train[col] = train[col].clip(lower, upper) elif method == "remove": train = train[(train[col] >= lower) & (train[col] <= upper)] else: raise ValueError("method must be 'clip' or 'remove'") return train COMMAND_BLOCK:
# Cap outliers print(f"Before outlier cleaning: {train_cleaned.shape}")
train_cleaned = handle_outliers(train_cleaned, num_cols, method="clip")
print(f"After outlier cleaning: {train_cleaned.shape}") Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Cap outliers print(f"Before outlier cleaning: {train_cleaned.shape}")
train_cleaned = handle_outliers(train_cleaned, num_cols, method="clip")
print(f"After outlier cleaning: {train_cleaned.shape}") COMMAND_BLOCK:
# Cap outliers print(f"Before outlier cleaning: {train_cleaned.shape}")
train_cleaned = handle_outliers(train_cleaned, num_cols, method="clip")
print(f"After outlier cleaning: {train_cleaned.shape}") COMMAND_BLOCK:
# Separate features and target
X= train_cleaned.drop("defects", axis=1)
y= train_cleaned["defects"]
X_train, X_test, y_train, y_test = train_test_split(X, y,test_size=0.1, random_state = 42, stratify=y) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Separate features and target
X= train_cleaned.drop("defects", axis=1)
y= train_cleaned["defects"]
X_train, X_test, y_train, y_test = train_test_split(X, y,test_size=0.1, random_state = 42, stratify=y) COMMAND_BLOCK:
# Separate features and target
X= train_cleaned.drop("defects", axis=1)
y= train_cleaned["defects"]
X_train, X_test, y_train, y_test = train_test_split(X, y,test_size=0.1, random_state = 42, stratify=y) COMMAND_BLOCK:
# Initialize Random Forest Classifier
rf_model = RandomForestClassifier( n_estimators=50, criterion='gini', max_depth=20, min_samples_split=10, min_samples_leaf=5, max_features='sqrt', bootstrap=True, random_state=42, n_jobs=-1
)
# Train the model
rf_model.fit(X_train, y_train)
print("Random Forest model trained successfully!") Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Initialize Random Forest Classifier
rf_model = RandomForestClassifier( n_estimators=50, criterion='gini', max_depth=20, min_samples_split=10, min_samples_leaf=5, max_features='sqrt', bootstrap=True, random_state=42, n_jobs=-1
)
# Train the model
rf_model.fit(X_train, y_train)
print("Random Forest model trained successfully!") COMMAND_BLOCK:
# Initialize Random Forest Classifier
rf_model = RandomForestClassifier( n_estimators=50, criterion='gini', max_depth=20, min_samples_split=10, min_samples_leaf=5, max_features='sqrt', bootstrap=True, random_state=42, n_jobs=-1
)
# Train the model
rf_model.fit(X_train, y_train)
print("Random Forest model trained successfully!") COMMAND_BLOCK:
# Display model parameters
print("\nModel Parameters:")
print(rf_model.get_params()) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Display model parameters
print("\nModel Parameters:")
print(rf_model.get_params()) COMMAND_BLOCK:
# Display model parameters
print("\nModel Parameters:")
print(rf_model.get_params()) COMMAND_BLOCK:
# Calculate accuracy scores
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred) print("=" * 50)
print("Accuracy Scores")
print("=" * 50)
print(f"Training Accuracy: {train_accuracy:.4f} ({train_accuracy*100:.2f}%)")
print(f"Testing Accuracy: {test_accuracy:.4f} ({test_accuracy*100:.2f}%)")
print("=" * 50) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Calculate accuracy scores
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred) print("=" * 50)
print("Accuracy Scores")
print("=" * 50)
print(f"Training Accuracy: {train_accuracy:.4f} ({train_accuracy*100:.2f}%)")
print(f"Testing Accuracy: {test_accuracy:.4f} ({test_accuracy*100:.2f}%)")
print("=" * 50) COMMAND_BLOCK:
# Calculate accuracy scores
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred) print("=" * 50)
print("Accuracy Scores")
print("=" * 50)
print(f"Training Accuracy: {train_accuracy:.4f} ({train_accuracy*100:.2f}%)")
print(f"Testing Accuracy: {test_accuracy:.4f} ({test_accuracy*100:.2f}%)")
print("=" * 50) COMMAND_BLOCK:
# Initialize Decision Tree Classifier
dt_model = DecisionTreeClassifier( criterion='gini', max_depth=5, min_samples_split=20, min_samples_leaf=10, random_state=42
) # Train the model
dt_model.fit(X_train, y_train) print("Decision Tree model trained successfully!") Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Initialize Decision Tree Classifier
dt_model = DecisionTreeClassifier( criterion='gini', max_depth=5, min_samples_split=20, min_samples_leaf=10, random_state=42
) # Train the model
dt_model.fit(X_train, y_train) print("Decision Tree model trained successfully!") COMMAND_BLOCK:
# Initialize Decision Tree Classifier
dt_model = DecisionTreeClassifier( criterion='gini', max_depth=5, min_samples_split=20, min_samples_leaf=10, random_state=42
) # Train the model
dt_model.fit(X_train, y_train) print("Decision Tree model trained successfully!") CODE_BLOCK:
from sklearn.tree import export_text tree_rules = export_text(dt_model)
print(tree_rules) Enter fullscreen mode Exit fullscreen mode CODE_BLOCK:
from sklearn.tree import export_text tree_rules = export_text(dt_model)
print(tree_rules) CODE_BLOCK:
from sklearn.tree import export_text tree_rules = export_text(dt_model)
print(tree_rules) COMMAND_BLOCK:
# Make predictions
y_train_pred = dt_model.predict(X_train)
y_test_pred = dt_model.predict(X_test) print("Predictions completed!") Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Make predictions
y_train_pred = dt_model.predict(X_train)
y_test_pred = dt_model.predict(X_test) print("Predictions completed!") COMMAND_BLOCK:
# Make predictions
y_train_pred = dt_model.predict(X_train)
y_test_pred = dt_model.predict(X_test) print("Predictions completed!") COMMAND_BLOCK:
# Calculate accuracy scores
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred) print("=" * 50)
print("Accuracy Scores")
print("=" * 50)
print(f"Training Accuracy: {train_accuracy:.4f} ({train_accuracy*100:.2f}%)")
print(f"Testing Accuracy: {test_accuracy:.4f} ({test_accuracy*100:.2f}%)")
print("=" * 50) Enter fullscreen mode Exit fullscreen mode COMMAND_BLOCK:
# Calculate accuracy scores
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred) print("=" * 50)
print("Accuracy Scores")
print("=" * 50)
print(f"Training Accuracy: {train_accuracy:.4f} ({train_accuracy*100:.2f}%)")
print(f"Testing Accuracy: {test_accuracy:.4f} ({test_accuracy*100:.2f}%)")
print("=" * 50) COMMAND_BLOCK:
# Calculate accuracy scores
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred) print("=" * 50)
print("Accuracy Scores")
print("=" * 50)
print(f"Training Accuracy: {train_accuracy:.4f} ({train_accuracy*100:.2f}%)")
print(f"Testing Accuracy: {test_accuracy:.4f} ({test_accuracy*100:.2f}%)")
print("=" * 50) CODE_BLOCK:
#linear Regression
def lr(X_train, X_test, y_train, y_test): model = LinearRegression() model.fit(X_train, y_train.values.ravel()) y_pred = model.predict(X_test) error = mean_squared_error(y_test, y_pred) print(f"Score: {error}") return model Enter fullscreen mode Exit fullscreen mode CODE_BLOCK:
#linear Regression
def lr(X_train, X_test, y_train, y_test): model = LinearRegression() model.fit(X_train, y_train.values.ravel()) y_pred = model.predict(X_test) error = mean_squared_error(y_test, y_pred) print(f"Score: {error}") return model CODE_BLOCK:
#linear Regression
def lr(X_train, X_test, y_train, y_test): model = LinearRegression() model.fit(X_train, y_train.values.ravel()) y_pred = model.predict(X_test) error = mean_squared_error(y_test, y_pred) print(f"Score: {error}") return model CODE_BLOCK:
#Submission
X_test = test[X_train.columns]
y_test_pred = rf_model.predict(X_test)
submission = pd.DataFrame({ "id": test["id"], "defects": y_test_pred
})
submission.to_csv("submission.csv", index=False)
submission Enter fullscreen mode Exit fullscreen mode CODE_BLOCK:
#Submission
X_test = test[X_train.columns]
y_test_pred = rf_model.predict(X_test)
submission = pd.DataFrame({ "id": test["id"], "defects": y_test_pred
})
submission.to_csv("submission.csv", index=False)
submission CODE_BLOCK:
#Submission
X_test = test[X_train.columns]
y_test_pred = rf_model.predict(X_test)
submission = pd.DataFrame({ "id": test["id"], "defects": y_test_pred
})
submission.to_csv("submission.csv", index=False)
submission
how-totutorialguidedev.toaimachine learningdockerpythongitgithub

